Rosetta @ Home (the origin)
A crowdsource computing project
Rosetta@home is a computing project of protein structure prediction that was designed an is being run by the University of Washington. Rosetta@home is a crowd source project that aims to predict protein-protein docking and desing new proteins. This is achieved by the volunteering of computers processing power around the world, achieving 487,946 GigaFLOPS on average as of September 19, 2020, though much of the project is oriented toward basic research to improve the accuracy and robustness of proteomics methods
Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes).
Rosetta@home uses idle computers processors, mainly focusing on volunteers computers who are not in use, to perform calculations on individual workunits. Completed calculations are sent to the main project server where once received are checked and validated to then be added into the main project’s database. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.
The Rosetta@home project consists of several front end web servers, primary/backup file servers, and master/slave MySQL servers. The network backbone between the servers and to the Internet runs at 10 Gbps (fiber and copper).
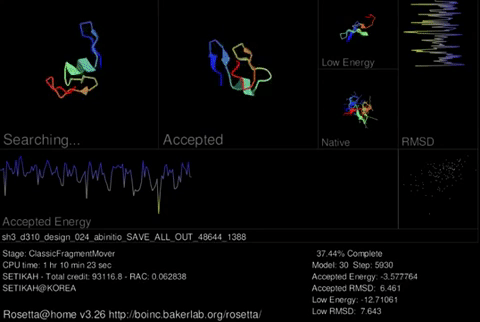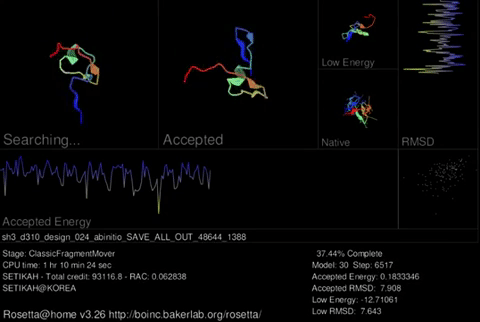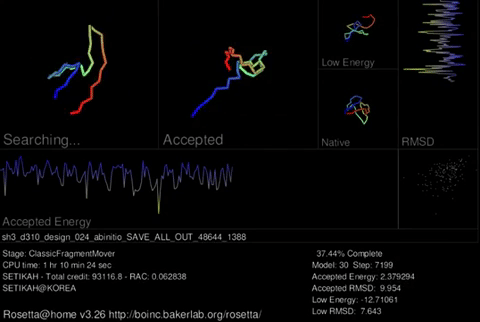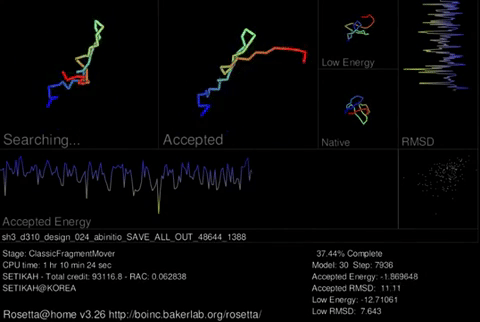
Rosetta@home uses two trajectories to predict the best protein structure. A trajectory may consist of two stages. The first stage uses a simplified representation of amino acids which allows us to try many different possible shapes rapidly. This stage is regarded as a low resolution search and on the screen saver you will see the protein chain jumping around a lot. In the second stage, Rosetta uses a full representation of amino acids. This stage is refered to as “relaxation.” Instead of moving around a lot, the protein tries smaller changes in an attempt to move the amino acids to their correct arrangment. This stage is regarded as a high resolution search and on the screen saver, you will see the protein chain jiggle around a little. Rosetta can do the first stage in a few minutes on a modern computer. The second stage takes longer because of the increased complexity when considering the full representation (all atoms) of amino acids.
The primary governing method of evaluation for rosetta@home CASP. CASP stands for Critical Assessment of protein Structure Prediction and is a community-wide, worldwide experiment for protein structure prediction taking place every two years since 1994. CASP provides research groups with an opportunity to objectively test their structure prediction methods and delivers an independent assessment of the state of the art in protein structure modeling to the research community and software users. (More on the next blog)
Rosetta@home is also used in immediate disease-related research, such fields that it has been involved have been:
- Alzheimer’s disease
- Anthrax
- Herpes simplex virus 1
- HIV
- Malaria
- COVID-19
- Cancer
Rosetta@Home and COVID-19
Rosetta has been used in molecular modelling to accurately predict the atomic-scale structure of the SARS-CoV-2 spike protein weeks before it could be measured in the lab. On June 26th of 2020, the project announced it had succeeded in creating antiviral proteins that neutralize SARS-CoV-2 virions(the complete, infective form of a virus outside a host cell) in the lab and that these experimental antiviral drugs are being optimized for animal testing trials.
In conclusion, Rosetta@Home can be used to put your idle computer processing power to good use and help current scientist fight the Covid pandemic or other diseases, visit Rosetta@Home’s webpage and download the screensaver now.
